2025
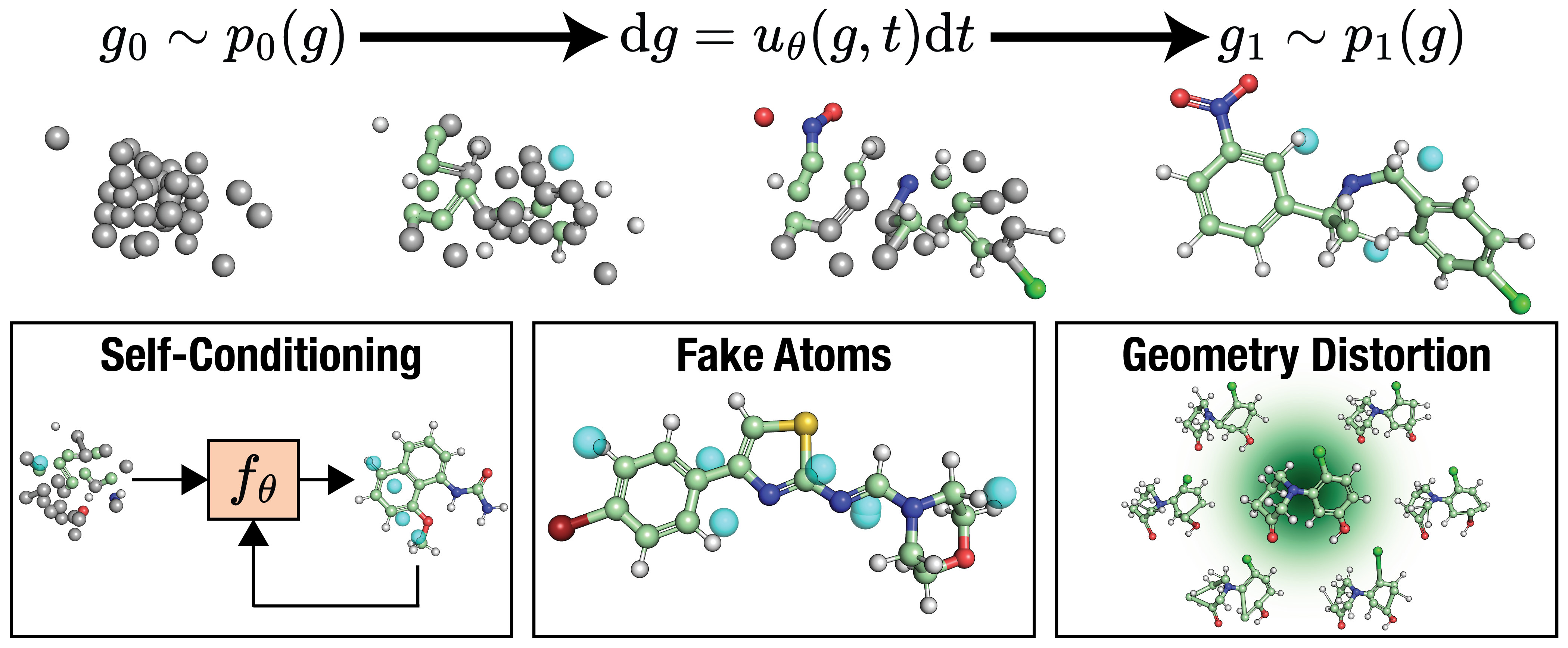
FlowMol3: Flow Matching for 3D De Novo Small-Molecule Generation
Ian Dunn, David Ryan Koes
Preprint September 2025
We present FlowMol3, a flow matching model that pushes the state of the art in unconditional 3D de novo small-molecule generation. FlowMol3 achieves nearly 100% molecular validity for drug-like molecules with explicit hydrogens, more accurately reproduces the functional group composition and geometry of its training data, and does so with an order of magnitude fewer learnable parameters than comparable methods.
FlowMol3: Flow Matching for 3D De Novo Small-Molecule Generation
Ian Dunn, David Ryan Koes
Preprint September 2025
We present FlowMol3, a flow matching model that pushes the state of the art in unconditional 3D de novo small-molecule generation. FlowMol3 achieves nearly 100% molecular validity for drug-like molecules with explicit hydrogens, more accurately reproduces the functional group composition and geometry of its training data, and does so with an order of magnitude fewer learnable parameters than comparable methods.
2024
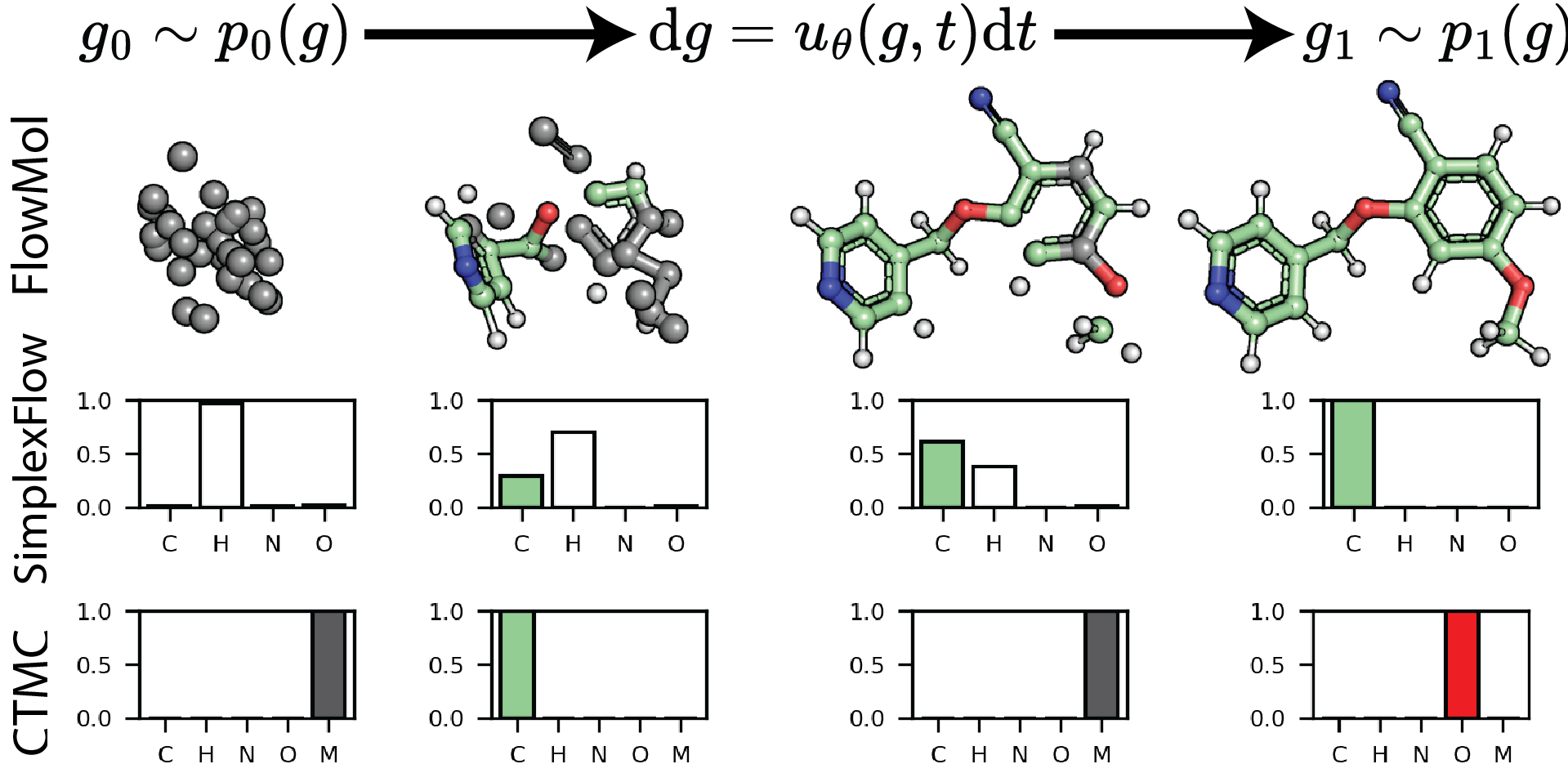
Exploring Discrete Flow Matching for 3D De Novo Molecule Generation
Ian Dunn, David Ryan Koes
NeurIPS 2024 Workshop on Machine Learning for Sturctural Biology November 2024
In this work we benchmark the performance of existing discrete flow matching methods for 3D de novo small molecule generation and provide explanations of their differing behavior. As a result we present FlowMol-CTMC, an open-source model that achieves state of the art performance for 3D de novo design with fewer learnable parameters than existing methods. Additionally, we propose the use of metrics that capture molecule quality beyond local chemical valency constraints and towards higher-order structural motifs.
Exploring Discrete Flow Matching for 3D De Novo Molecule Generation
Ian Dunn, David Ryan Koes
NeurIPS 2024 Workshop on Machine Learning for Sturctural Biology November 2024
In this work we benchmark the performance of existing discrete flow matching methods for 3D de novo small molecule generation and provide explanations of their differing behavior. As a result we present FlowMol-CTMC, an open-source model that achieves state of the art performance for 3D de novo design with fewer learnable parameters than existing methods. Additionally, we propose the use of metrics that capture molecule quality beyond local chemical valency constraints and towards higher-order structural motifs.
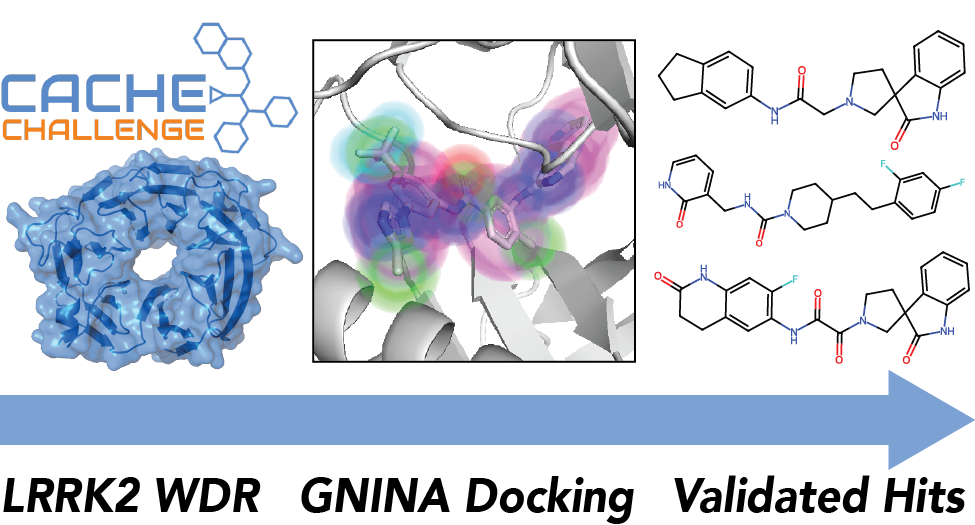
CACHE Challenge #1: Docking with GNINA is all you need
Ian Dunn, Somayeh Pirhadi, Yao Wang, Smmrithi Ravindran, Carter Concepcion, David Ryan Koes
Journal of Chemical Information and Modeling December 2024
We describe our winning submission to the first Critical Assessment of Computational Hit-Finding Experiments (CACHE) challenge. Our screening campaign was largely built around gnina, an open-source molecular docking program developed by the Koes lab that uses a deep-learning based scoring function. Our resulting best hit series tied for first place when evaluated by a panel of expert judges.
CACHE Challenge #1: Docking with GNINA is all you need
Ian Dunn, Somayeh Pirhadi, Yao Wang, Smmrithi Ravindran, Carter Concepcion, David Ryan Koes
Journal of Chemical Information and Modeling December 2024
We describe our winning submission to the first Critical Assessment of Computational Hit-Finding Experiments (CACHE) challenge. Our screening campaign was largely built around gnina, an open-source molecular docking program developed by the Koes lab that uses a deep-learning based scoring function. Our resulting best hit series tied for first place when evaluated by a panel of expert judges.
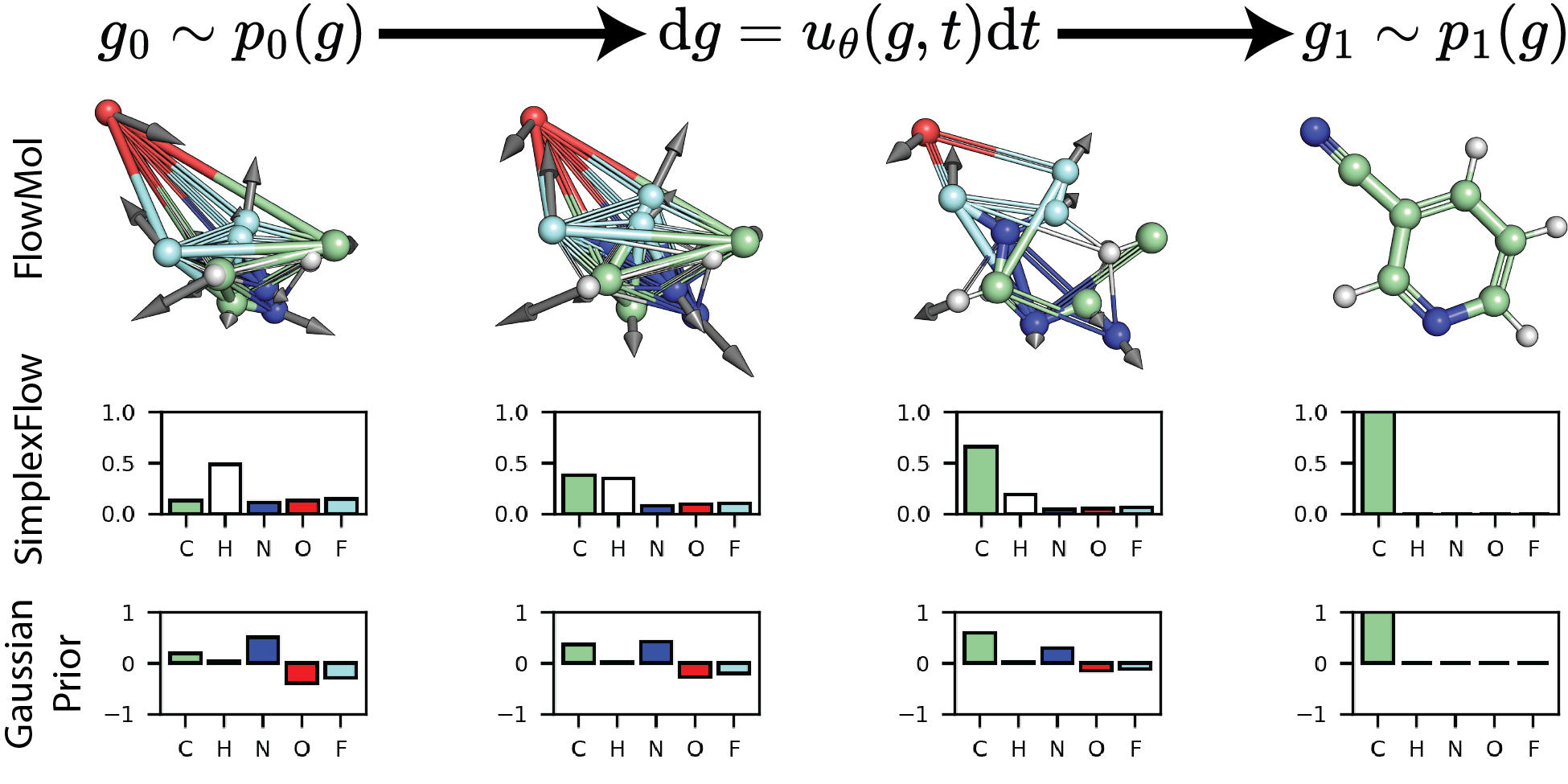
Mixed Continuous and Categorical Flow Matching for 3D De Novo Molecule Generation
Ian Dunn, David Ryan Koes
Preprint April 2024
We develop FlowMol: the first flow matching model that samples the geometric and topological structure of organic molecules. We also develop a method for flow matching on discrete data that we call SimplexFlow.
Mixed Continuous and Categorical Flow Matching for 3D De Novo Molecule Generation
Ian Dunn, David Ryan Koes
Preprint April 2024
We develop FlowMol: the first flow matching model that samples the geometric and topological structure of organic molecules. We also develop a method for flow matching on discrete data that we call SimplexFlow.
2023
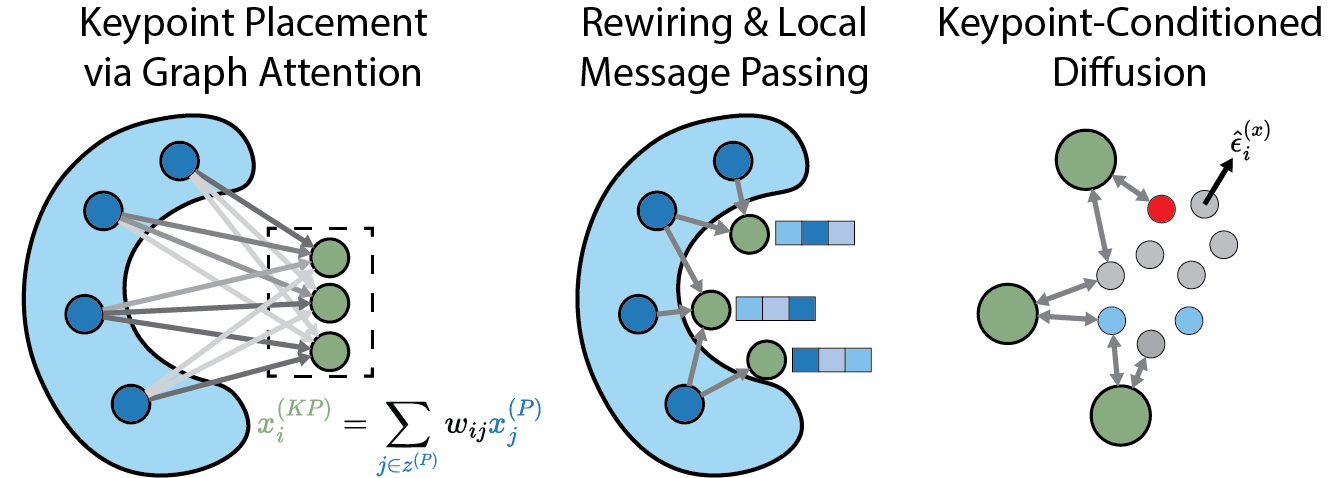
Accelerating Inference in Molecular Diffusion Models with Latent Representations of Protein Structure
Ian Dunn, David Ryan Koes
NeurIPS 2024 Workshop on Generative AI for Biology October 2023 Spotlight
We present a novel method for learning compressed geometric representations of protein structure and use this novel protein encoder to accelerate inference in diffusion models.
Accelerating Inference in Molecular Diffusion Models with Latent Representations of Protein Structure
Ian Dunn, David Ryan Koes
NeurIPS 2024 Workshop on Generative AI for Biology October 2023 Spotlight
We present a novel method for learning compressed geometric representations of protein structure and use this novel protein encoder to accelerate inference in diffusion models.